Expressing differences in morphology
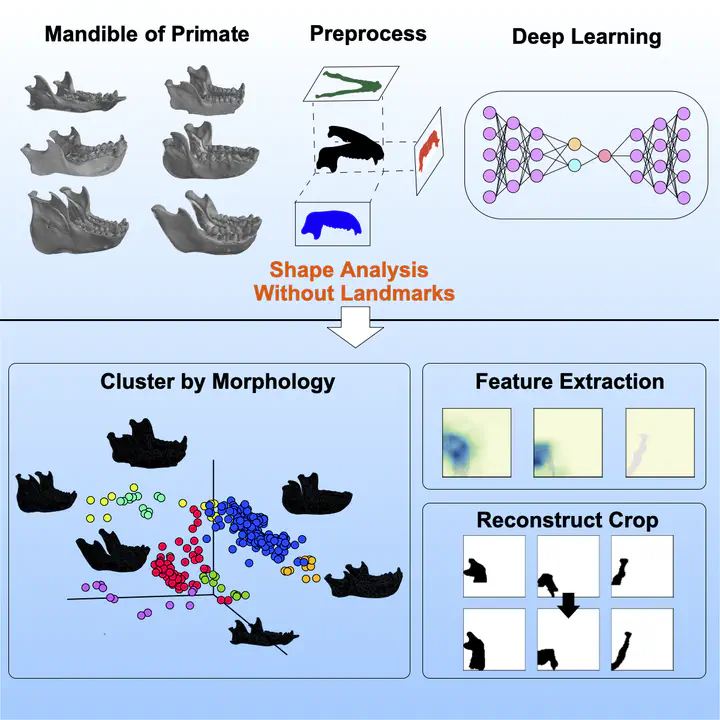
Shape measurement is critical to evolutionary and developmental biology, but it is difficult to objectively and automatically quantify arbitrary shapes. Traditional approaches are based on anatomically salient landmarks and require manual annotation by experts.
Therefore, I developed an image-based deep learning framework, Morphological regulated variational AutoEncoder (Morpho-VAE). This is a machine learning approach for landmark-free shape analysis. The proposed architecture combines unsupervised and supervised learning models and reduces dimensionality by focusing on morphological features that distinguish data with different labels. We applied this approach to primate mandible image data. Despite the lack of correlation between the extracted morphological features and phylogenetic distance, the extracted morphological features reflected the characteristics of the family to which the organism belonged. Furthermore, it was shown that the missing parts can be recovered from incomplete images. The proposed method provides a flexible and promising analysis tool for various image data of biological shapes, even for image data containing missing segments.
